OSU, OU scientists collaborate to sequence, map new genome
Thursday, February 10, 2022
Media Contact: Jacob Longan | Coordinator of Communications and Marketing, College of Arts and Sciences | 405-744-7497 | jacob.longan@okstate.edu
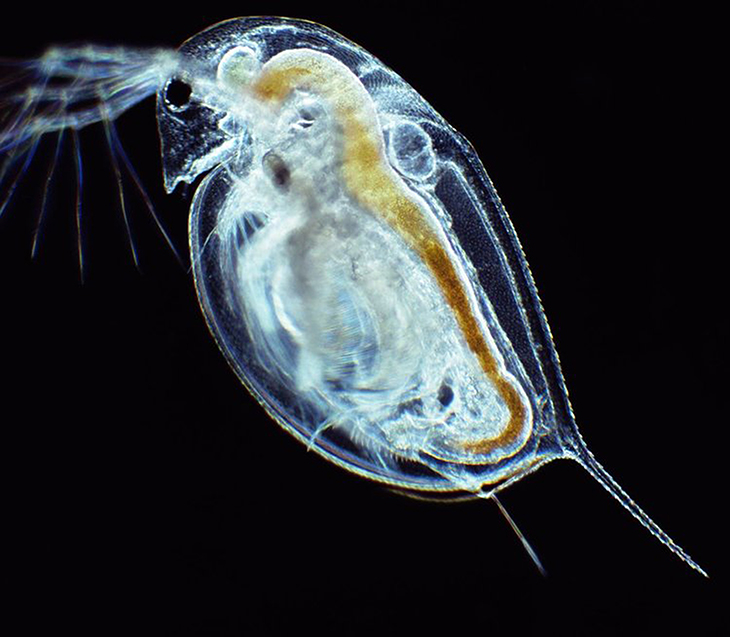
A team of researchers from Oklahoma State University and the University of Oklahoma published a genome sequence and chromosome map for the micro-crustacean Daphnia pulicaria.
Researchers in the laboratory of Punidan Jeyasingh in OSU’s Department of Integrative Biology — in collaboration with the laboratory of OU biology professor Lawrence Weider — led the efforts. The results of their work were published in the journal Molecular Ecology, as part of a special issue highlighting the latest advances in DNA sequencing technology.
Daphnia, also known as waterfleas, play critical roles in environmental monitoring. For example, they have served as an important test organism in laboratories around the world for over a century because of their sensitivity to many environmental stressors such as chemicals. In nature, Daphnia act as a keystone species in freshwater food webs globally, as they feed on algae to help keep lake, pond and reservoir water clear and are important food sources for recreational fish species.
The researchers used HiFi DNA sequencing technology, which produces highly accurate and very long stretches of DNA “letters” known as sequencing reads.
“Many people describe assembling a genome as a bit like putting together a huge jigsaw puzzle, fitting together many smaller pieces to form a whole image,” said Matthew Wersebe, a doctoral candidate at OU and lead author of the study. “When the pieces are large and the picture crisp, the whole process is a lot easier.”
Jeyasingh added, “These technological advances are answering longstanding questions in ecology and evolutionary biology, and I’m glad the next generation, like Matt, are mastering these methods and advancing our science.”
The use of this type of DNA sequencing technology allowed the researchers to piece together the genome into huge and continuous stretches of DNA and also discern the two copies of the genome, one belonging to each of the two parents of the Daphnia individual they sequenced. To map and orient the long DNA chunks into chromosomes, the researchers also used genome sequencing of 100 “recombinant” individuals that they bred in the lab as part of the project. Finally, the researchers compared their new genome of Daphnia pulicaria to other closely related species. They determined that because Daphnia pulicaria has recently colonized lakes, it has experienced significant natural selection in its recent history as it has adapted to its new lake habitat. The new high-quality genome presented in the study allowed the researchers to examine the genomic basis of speciation in Daphnia.
The genome sequence and chromosome map will form the basis of future collaborations between the Jeyasingh and Weider labs, where researchers are studying how waterfleas respond to changes in their environment from stressors like nutrients and road salt pollution, to invasive species and climate change.
The work was funded in part by the National Science Foundation and the OU Board of Regents.
For more information, contact Matthew Wersebe at matthew.wersebe@ou.edu.
