COVID-19 surge prediction now possible with genomic surveillance
Thursday, February 2, 2023
Media Contact: Kaylie Wehr | OSU College of Veterinary Medicine Communications Coordinator | 405-744-6740 | kaylie.wehr@okstate.edu
Using genomic surveillance, Oklahoma State University researchers have developed a new method of predicting infectious disease surges, including COVID-19.
Preparation time is crucial for prevention and something medical professionals lacked at the start of the COVID-19 pandemic. That spurred Dr. Pratul Agarwal, assistant vice president of research and professor of physiological sciences in the OSU College of Veterinary Medicine, and his colleagues, to explore ways to predict waves of increased infections, or so-called surges, ahead of time by analyzing data collected through genome sequencing, which is the process of deciphering the genetic material found in the virus. Agarwal’s team recently published its findings in the prestigious eLife, a peer-reviewed, scientific journal for biomedical and life sciences. Full details are available at https://elifesciences.org/articles/82980.
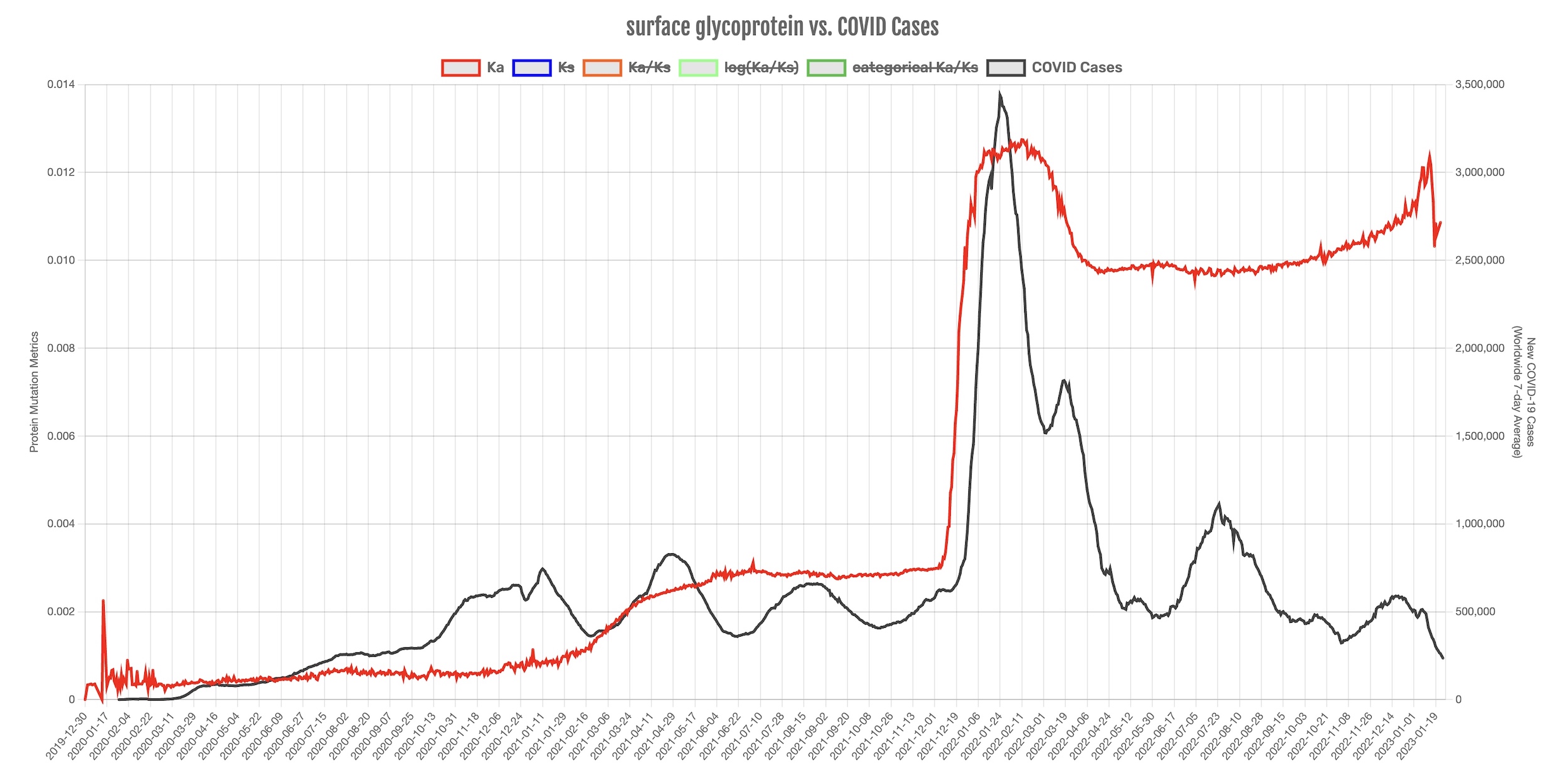
“The availability of millions of SARS-CoV-2 genomic sequences was a silver lining on the very dark cloud of the pandemic. Nothing had been done at this scale before,” Agarwal said. “A set of samples from COVID-19 positive patients are regularly being sequenced to see what the virus is doing. Over 6 million sequences are now available through public databases such as the GenBank of National Center for Biotechnology Information (NCBI). We thought, maybe we could use this data in real-time as it becomes available to see how fast the virus is mutating and what it could do next. And if we get it right, then this information can then help medical professionals prepare for increased cases.”
The team combined their skills in bioinformatics, data analysis and software engineering to put together a framework for real-time genomic surveillance. They collaborated with experts from Dr. Shozeb Haider’s team at the University College London School of Pharmacy to make this possible. The resulting information is now being shared in the form of a pandemic preparedness dashboard and alert system aimed at providing medical personnel and the general public time needed to prepare for an influx of infectious disease cases.
This framework is updated daily and is available to the research and medical community, as well as the general public, athttp://pandemics.okstate.edu/covid19/. Anyone can sign up to receive updates directly to their email inbox.
As a result, Agarwal’s team can now predict COVID-19 surges two to four weeks before they occur and have successfully predicted every surges since fall of last year, including the ones associated with the Omicron and related SARS-CoV-2 variants.
“Our scientists are fighting against the next pandemic right now,” said Oklahoma Secretary of Science and Innovation Elizabeth Pollard. “What Dr. Agarwal’s team has done is to create a tool that allows us to prepare for future pandemics. Such tools will help our medical community and save lives in the future.”
In addition to assisting medical professionals, these predictions can also be helpful to the general public.
When a potential surge is identified, the dashboard indicates the potential of a surge in infection cases by issuing watches and warnings. A watch indicates a potential for a surge whereas a warning indicates a high confidence that a surge will occur.
So how does it work exactly?
“The virus has 26 proteins that do different things and many of us have already heard about the spike protein, which is the protein that is a key player in infecting the human cells,” Agarwal said. “Our study found that the spike protein is heavily mutating in its search for entering the human cells. And that’s just an example of one protein. Many of the other proteins are also mutating to evade the machinery of the human immune system.”
The team is now able to look through the large amounts of sequences that come through the databases and keep an eye on the mutations. This process is known as genomic surveillance. When researchers begin to notice a rise in certain mutations in several samples, they use the algorithm they developed to collect information and make a prediction on which mutations will have a high probability of increasing the infection rate.
“If the mutation rates start to change rapidly, we issue an alert,” Agarwal said. “We saw that on several occasions, the mutations in the next 10 to 14 days cause a large number of infections. So, we try to catch important mutations before they cause a chain reaction of cases. Some feel that COVID-19 is in the rear-view mirror, but viruses are always mutating. We need to prepare in case this virus mutates heavily again so much that current vaccines and antiviral drugs become less effective or even ineffective. Keeping an eye on mutations in real-time could help in keeping diagnostic kits up to date and designing new treatments.”
Now that all the pieces are in place, this system could even be used to make predictions about future infectious diseases, such as monkeypox or possibly even the flu if sufficient genomic sequences become available, making it an important piece of the future pandemic prevention strategy.
